Walkthrough
Learn how to get the most out of ACME for your own work by running through a quick but (hopefully) illustrative example that starts simple and subsequently turns on ACME’s bells and whistles.
Setup
Assume the function defined below is supposed to be run multiple times
in parallel for different values of x, y and z
def f(x, y, z=3):
return (x + y) * z
The following code calls f with four different values of x (namely 2, 4, 6 and 8)
sets y to 4 (for all calls) and similarly leaves z at its default value of 3:
from acme import ParallelMap
with ParallelMap(f, [2, 4, 6, 8], 4) as pmap:
results = pmap.compute()
What’s going on here? ACME starts a dask distributed.Client containing
four parallel workers (or fewer depending on available resources) that
call f with the provided input values to perform the following calculations
concurrently:
Computation #0: 18 = (2 + 4) * 3 (
x = 2,y = 4,z = 3)Computation #1: 24 = (4 + 4) * 3 (
x = 4,y = 4,z = 3)Computation #2: 30 = (6 + 4) * 3 (
x = 6,y = 4,z = 3)Computation #3: 36 = (8 + 4) * 3 (
x = 8,y = 4,z = 3)
Where Are My Results?
By default results are saved to disk in HDF5 format and results is a list
of the corresponding filenames:
>>> results
['/my/current/workdir/ACME_20221007-100302-976973/f_payload/f_0.h5',
'/my/current/workdir/ACME_20221007-100302-976973/f_payload/f_1.h5',
'/my/current/workdir/ACME_20221007-100302-976973/f_payload/f_2.h5',
'/my/current/workdir/ACME_20221007-100302-976973/f_payload/f_3.h5']
Note that ACME auto-generates a directory named ACME_YYYYMMDD-hhmmss-ffffff
(encoding the current time as YearMonthDay-HourMinuteSecond-Microsecond).
Each evaluation of f is stored in a dedicated HDF5 file, that in turn is
referenced in an aggregate results container pmap.results_container
(comprised of external links):
>>> pmap.results_container
'/my/current/workdir/ACME_20221007-100302-976973/f.h5'
In the above example, four HDF5 files are generated: f_0.h5, f_1.h5,
f_2.h5, f_3.h5, each comprising the result of calling f with the provided
input values (i.e., f_0.h5 stores the result of Computation #0: 18 = (2 + 4) * 3,
f_1.h5 holds the result of Computation #1: 24 = (4 + 4) * 3 and so on).
The collective results of all calls of f are gathered in a single HDF5
container f.h5 whose group-/data-set structure follows the format shown below:
comp_0
└─ result_0
comp_1
└─ result_0
comp_2
└─ result_0
comp_3
└─ result_0
The contents of the containers can be accessed using h5py:
import h5py
with h5py.File(pmap.results_container, "r") as h5f:
comp0 = h5f["comp_0"]["result_0"][()]
comp1 = h5f["comp_1"]["result_0"][()]
comp2 = h5f["comp_2"]["result_0"][()]
comp3 = h5f["comp_3"]["result_0"][()]
print(comp0, comp1, comp2, comp3)
>>> [18] [24] [30] [36]
The dataset name “result_0” stores the first return value of the
user-provided function f. If f returns multiple quantities, analogously named
datasets ‘result_0’, “result_1”, “result_2”, … are automatically created
in the constructed HDF5 containers. See Auto-Generated HDF5-Files for more details.
Note
By default, the aggregate results container generated by ACME does not
contain actual data but only points to the corresponding HDF5 files
found in the respective _payload directory. Thus, the results container
is only meaningful in conjunction with its associated payload.
To override the target directory ACME writes HDF5 containers to, the
output_dir keyword can be provided:
from acme import ParallelMap
with ParallelMap(f, [2, 4, 6, 8], 4, output_dir="/path/to/folder") as pmap:
results = pmap.compute()
Then
>>> results
['/path/to/folder/f_payload/f_0.h5',
'/path/to/folder/f_payload/f_1.h5',
'/path/to/folder/f_payload/f_2.h5',
'/path/to/folder/f_payload/f_3.h5']
and
>>> pmap.results_container
'/path/to/folder/f.h5'
Note
While ACME’s default storage format is HDF5, user-functions that return
non-HDF compatible objects can be processed too as long as the returned
quantities are serializable. By setting write_pickle to True
when calling ParallelMap, ACME pickles results instead
of creating HDF5 containers. See Alternative Storage Format: Pickle (write_pickle) for an example and more
information. In addition, ACME uses an “emergency pickling” strategy to
save results if at all possible: if the output of some computational runs
cannot be stored in HDF5, ACME switches to on-demand pickling regardless
of the provided write_pickle setting.
Alternatively, results may be collected directly in memory by setting
write_worker_results to False. This is not recommended, since
values have to be gathered from compute nodes via ethernet (slow) and
are accumulated in the local memory of the interactive node you are using
(potential memory overflow):
with ParallelMap(f, [2, 4, 6, 8], 4, write_worker_results=False) as pmap:
results = pmap.compute()
Now results is a list of integers:
>>> results
[18, 24, 30, 36]
Override Automatic Input Argument Distribution
Next, suppose f has to be evaluated for the same values of x (again
2, 4, 6 and 8), but y is not a number but a NumPy array:
import numpy as np
y = np.ones((3,)) * 4
with ParallelMap(f, [2, 4, 6, 8], y) as pmap:
results = pmap.compute()
This fails, because it is not clear which input is to be split up and distributed across workers for parallel execution:
ValueError: <ParallelMap> automatic input distribution failed: found 2 objects containing 3 to 4 elements. Please specify n_inputs manually.
In this case, n_inputs has to be provided explicitly (write_worker_results
is set to False for illustration purposes only)
with ParallelMap(f, [2, 4, 6, 8], y, n_inputs=4, write_worker_results=False) as pmap:
results = pmap.compute()
yielding
>>> results
[array([18., 18., 18.]),
array([24., 24., 24.]),
array([30., 30., 30.]),
array([36., 36., 36.])]
Note that setting n_inputs manually can also be used to execute a function
n_inputs times with the same arguments (again write_worker_results
is set to False for illustration purposes only):
with ParallelMap(f, 2, 3, n_inputs=4, write_worker_results=False) as pmap:
results = pmap.compute()
Then
>>> results
[15, 15, 15, 15]
This functionality is sometimes useful for routines that randomize their in- and/or outputs. An example and more information is provided in Randomization and Concurrency (taskID)
Collect Results in Single Dataset
When evaluating functions that return a NumPy array (like in the example above),
it is sometimes beneficial to aggregate results in a single dataset. Assume
the four 1d-arrays of the above parallel evaluation of f are to be
collected in a single 2d-array. This can be achieved by specifying the keyword
result_shape where a (single!) None entry delineates the “stacking dimension”
along which computed results are to be put together
y = np.ones((3,)) * 4
with ParallelMap(f, [2, 4, 6, 8], y, n_inputs=4, result_shape=(None, 3)) as pmap:
results = pmap.compute()
The generated container is structured as follows:
>>> h5f = h5py.File(pmap.results_container, "r")
>>> h5f.keys()
<KeysViewHDF5 ['result_0']>
>>> h5f["result_0"]
<HDF5 dataset "result_0": shape (4, 3), type "<f8">
>>> h5f["result_0"][()]
array([[18., 18., 18.],
[24., 24., 24.],
[30., 30., 30.],
[36., 36., 36.]])
Instead of four HDF5 groups (“comp_0”, …, “comp_3”) each containing one
dataset (“result_0”), only a single dataset “result_0” is generated, whose
dimension is set a-priori via result_shape = (None, 3): this
tells ACME that incoming results are 3-component vectors, that are to be stacked
along the first dimension (position of None) of a 2d-dataset.
Similarly, results may be collected in memory by setting write_worker_results
to False (not recommended):
y = np.ones((3,)) * 4
with ParallelMap(f, [2, 4, 6, 8], y, n_inputs=4, result_shape=(None, 3), write_worker_results=False) as pmap:
results = pmap.compute()
This yields:
>>> results
[array([[18., 18., 18.],
[24., 24., 24.],
[30., 30., 30.],
[36., 36., 36.]])]
Note that in contrast to the example given in the previous section
Override Automatic Input Argument Distribution, results does not
contain four (3,)-arrays, but one (4, 3)-array. More information and additional
control options are discussed in Advanced Usage and Customization.
Reuse Worker Clients
Instead of letting ACME automatically start and stop parallel worker clients
witch each invocation of ParallelMap, a dask distributed.Client
can be customized and set up manually before launching the actual concurrent
computation. The convenience functions slurm_cluster_setup() (on HPC
clusters managed by the
SLURM Workload Manager)
and local_cluster_setup() (on local multi-core machines) provide
this functionality by wrapping dask_jobqueue.SLURMCluster and
distributed.LocalCluster, respectively. Once a client has been set up,
any subsequent invocation of ParallelMap automatically picks
up the allocated client and distributes computational payload across the
workers collected inside.
Note
The routine esi_cluster_setup() is specifically geared to the
SLURM setup of the ESI HPC cluster. If you are working on the ESI cluster,
please use esi_cluster_setup() to allocate computing clients.
Alternatively, instead of manually setting up computing resources using the
*_cluster_setup routines, any distributed client automatically sized and
started by ParallelMap can be re-used for subsequent
computations by setting the stop_client keyword to False.
Assume ACME is used on a HPC cluster managed by SLURM and suppose f
needs to be evaluated for fixed values of x and y with z varying randomly 500 times between 1 and 10. Since f is a
very simple function, it is not necessary to spawn 500 SLURM workers (=jobs) for this.
Instead, allocate only 50 workers in the “smallest” available queue on your
cluster (“8GBXS” on the ESI HPC cluster), i.e., each worker has to perform
10 evaluations of f. Additionally, keep the workers alive for re-use afterwards
x = 2
y = 4
rng = np.random.default_rng()
z = rng.integers(low=1, high=10, size=500, endpoint=True)
with ParallelMap(f, x, y, z=z, n_workers=50, partition="8GBXS", stop_client=False) as pmap:
results = pmap.compute()
This yields
>>> len(results)
500
In a subsequent computation f needs to be evaluated for 1000 samples of
z. In the previous call, stop_client was False, thus the next
invocation of ParallelMap re-uses the allocated distributed.Client
object containing 50 SLURM workers:
z = rng.integers(low=1, high=10, size=1000, endpoint=True)
with ParallelMap(f, x, y, z=z) as pmap:
results = pmap.compute()
Note the info message:
<ParallelMap> INFO: Attaching to global parallel computing client <Client: 'tcp://10.100.32.5:39747' processes=50 threads=50, memory=400.00 GB>
Debugging And Estimating Resource Consumption
Debugging programs running in parallel can be quite tricky.
For instance, assume the function f is (erroneously) called with z
set to None. In a regular sequential setting, identifying the problem
is (relatively) straight-forward:
>>> f(2, 4, z=None)
TypeError: unsupported operand type(s) for *: 'int' and 'NoneType'
However, when executing f in parallel using SLURM
with ParallelMap(f, [2, 4, 6, 8], 4, z=None) as pmap:
results = pmap.compute()
the resulting error message can be somewhat overwhelming
Function: execute_task
args: ((<function reify at 0x7f425c25b0d0>, (<function map_chunk at 0x7f425c25b4c0>,
<function ACMEdaemon.func_wrapper at 0x7f42569f1e50>, [[2], [4], [None], ['/cs/home/fuertingers/ACME_20201217-160137-984430'],
['f_0.h5'], [0], [<function f at 0x7f425c34bee0>]], ['z', 'outDir', 'outFile', 'taskID', 'userFunc'], {})))
kwargs: {}
Exception: TypeError("unsupported operand type(s) for *: 'int' and 'NoneType'")
slurmstepd: error: *** JOB 1873974 ON esi-svhpc18 CANCELLED AT 2020-12-17T16:01:43 ***
To narrow down problems with parallel execution, the compute()
method of ParallelMap offers the debug keyword. If enabled, all function calls
are performed in the local thread of the active Python interpreter. Thus, the actual execution
is not performed in parallel. This allows regular error propagation
and even permits the use of tools like pdb
or %debug iPython magics.
with ParallelMap(f, [2, 4, 6, 8], 4, z=None) as pmap:
results = pmap.compute(debug=True)
which results in
<ipython-input-2-47feb885f020> in f(x, y, z)
1 def f(x, y, z=3):
----> 2 return (x + y) * z
TypeError: unsupported operand type(s) for *: 'int' and 'NoneType'
In addition, ACME can be used to estimate memory consumption as well as runtime of compute jobs before actually launching a full concurrent processing run. This functionality permits to get a (rough) estimate of resource requirements for queuing systems and it allows to test-drive ACME’s automatically generated argument lists prior to the actual concurrent computation. For instance,
>>> with ParallelMap(f, [2, 4, 6, 8], 4, dryrun=True) as pmap:
>>> results = pmap.compute()
<ParallelMap> INFO: Performing a single dry-run of f simulating randomly picked worker #1 with automatically distributed arguments
<ParallelMap> INFO: Dry-run completed. Elapsed time is 0.004725 seconds, estimated memory consumption was 0.01 MB.
Do you want to continue executing f with the provided arguments? [Y/n] n
In general it is strongly recommended to make sure any function supplied
to ParallelMap works as intended in a sequential setting prior to running
it in parallel.
Interactive Monitoring
When ACME starts a distributed.Client, dask automatically sets up
a diagnostic dashboard
for the client. The dashboard is a web interface that allows live monitoring
of workers and their respective computations. ACME displays the link
for connecting to the dashboard as soon as it successfully launched a new
distributed computing client. For instance, invoking ParallelMap
on a local machine prints:
<local_cluster_setup> Cluster dashboard accessible at http://127.0.0.1:8787/status
Clicking on the link (or copy-pasting it to your browser) opens the client’s diagnostic
dashboard. This web interface offers various ways to monitor the current
state, memory and CPU usage of parallel workers and also provides an overview
of the global status of the concurrent processing task started by ParallelMap:
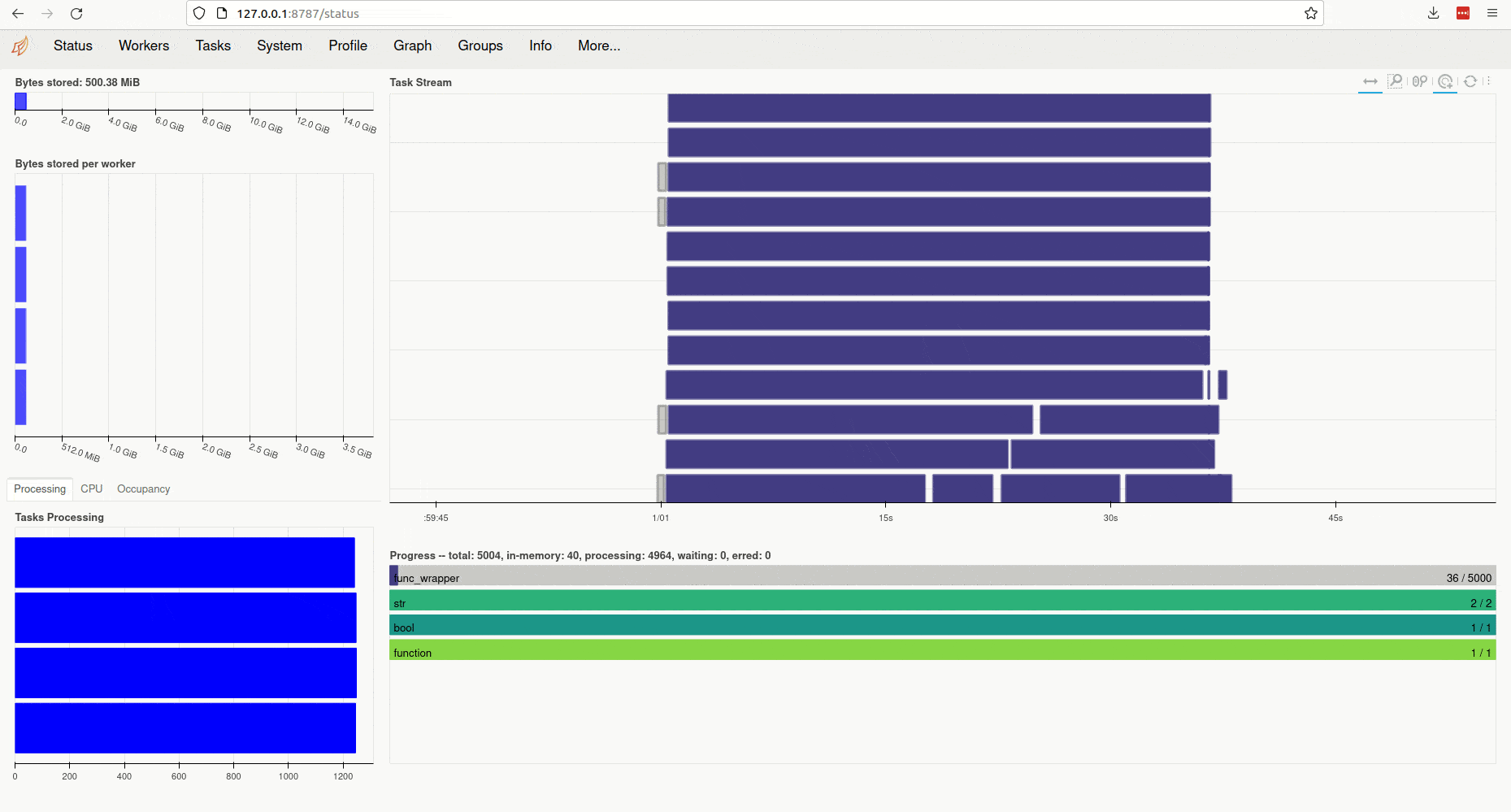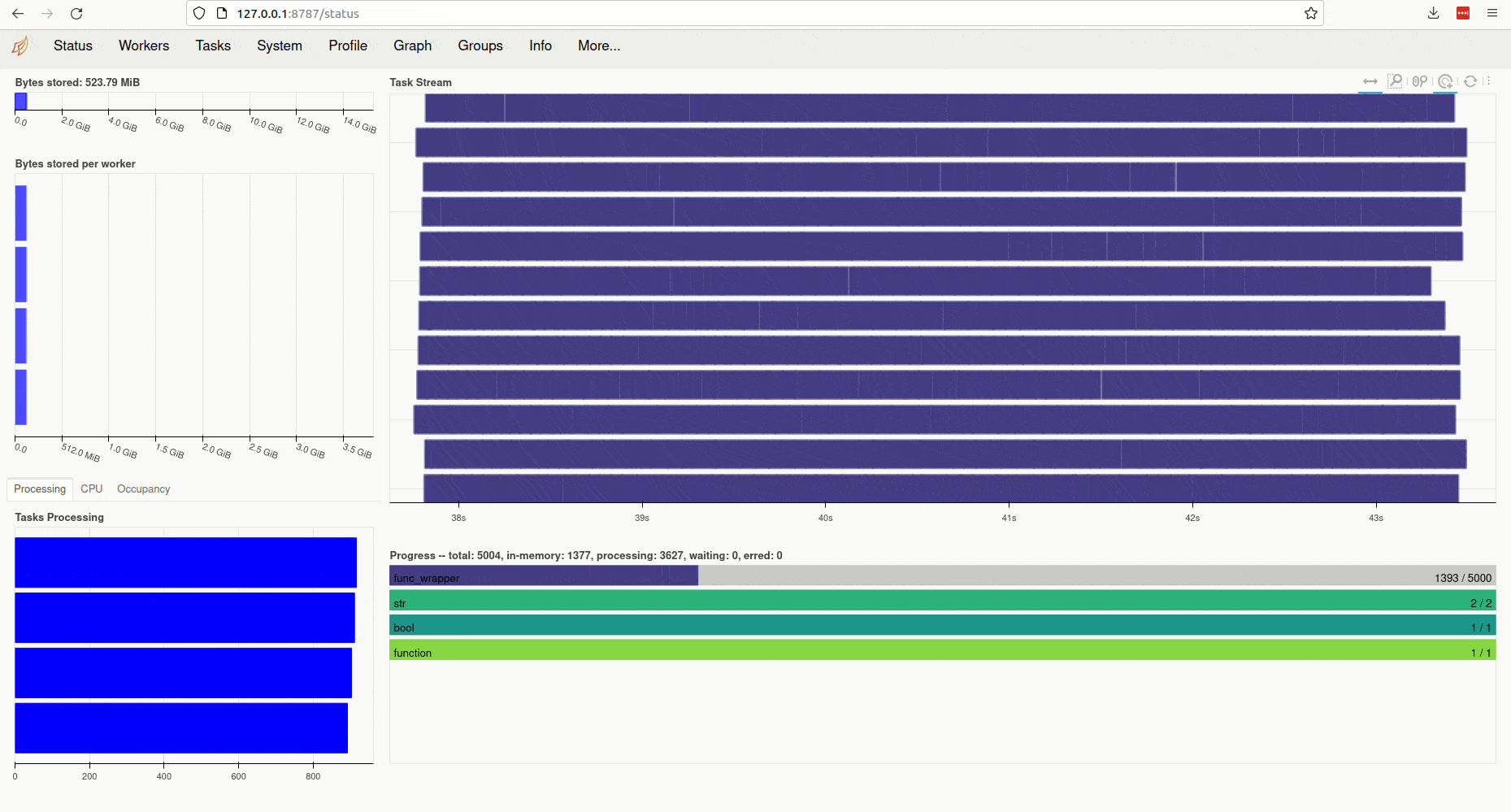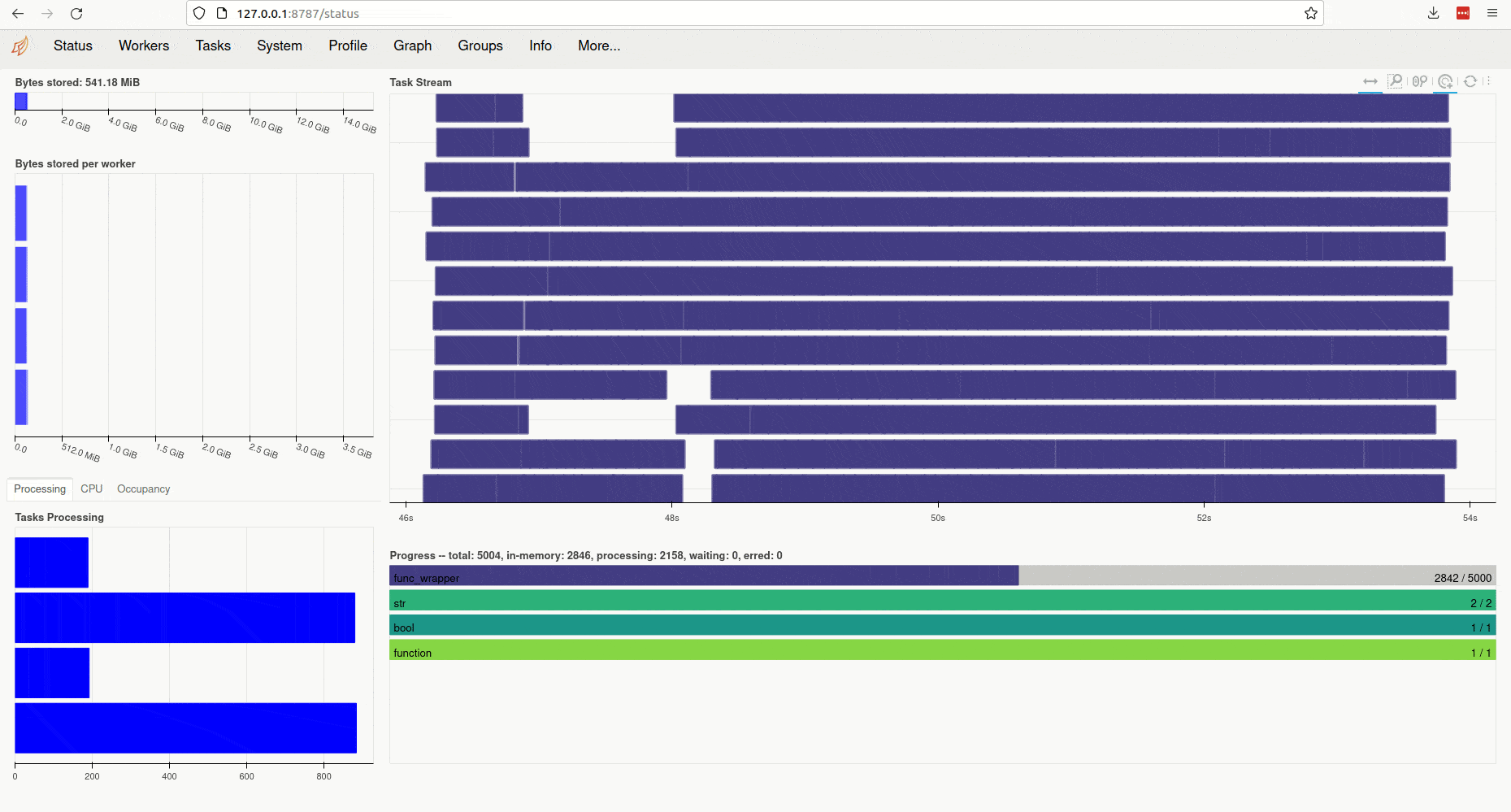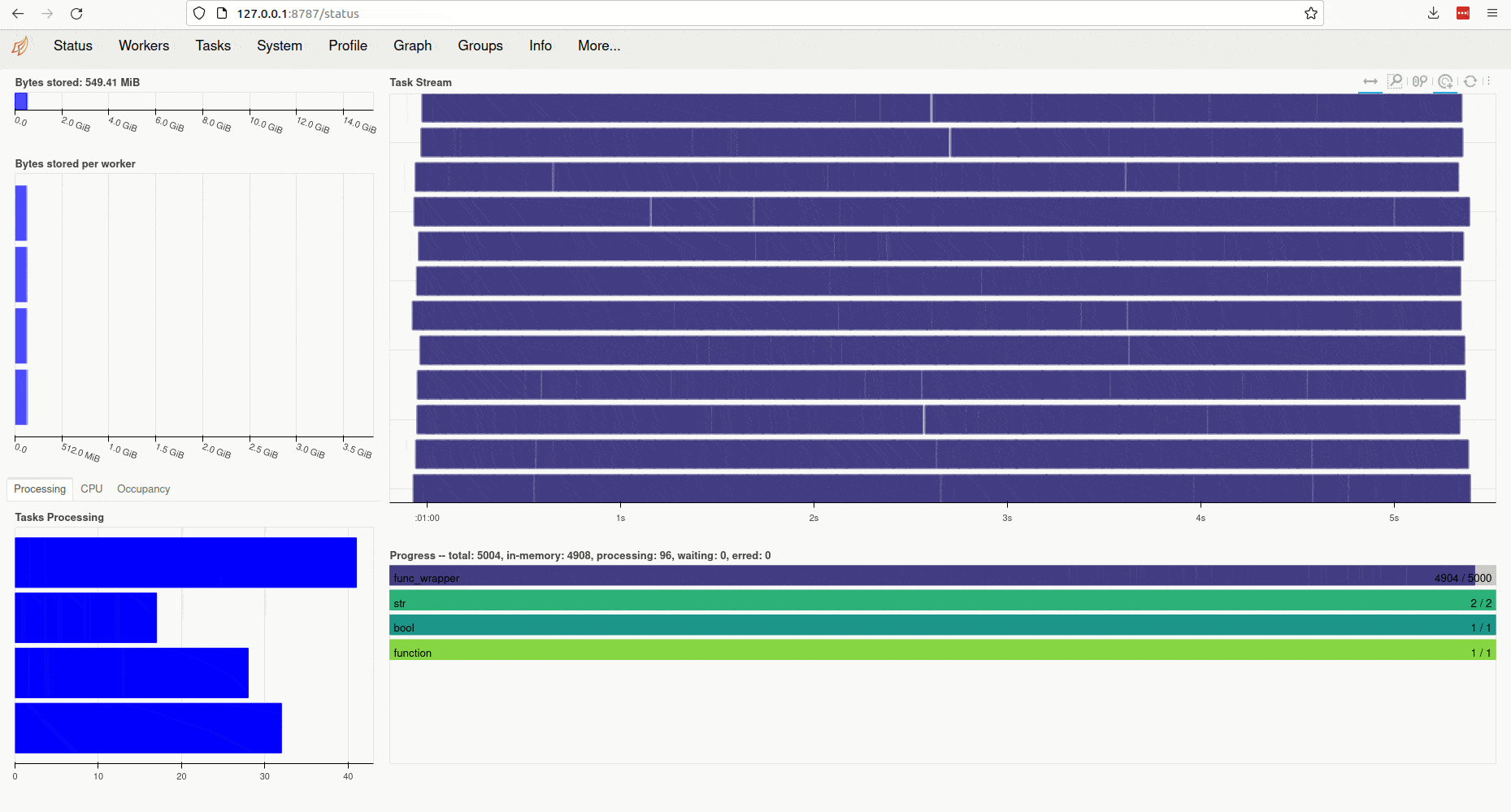
Non-Interactive Logging
In addition, if ACME’s automatic result management is used (i.e., if
write_worker_results is True), a log-file is created alongside
the files generated on disk. Depending on the chosen verbosity level of
ACME’s messaging system (i.e., if verbose is either False, None or
True), the generated log-file contains (very) detailed runtime
information of the performed computation.
Wait, There’s More…
ACME attempts to be as agnostic of user-provided functions as possible. However,
there are some technical limitations that impose medium to hard boundaries as to
what a user-provided function func can and should do. Most important, input
arguments of func must be regular Python objects (lists, tuples, scalars,
strings, etc.) or NumPy arrays. If ACME’s HDF5 storage backend is used, then
additionally func’s return values must be HDF5 compatible (i.e., scalars,
arrays or strings). More information and technical background is provided in
Advanced Usage and Customization.
