Parallel Bootstrap Tutorial
Note
These examples were run on the ESI HPC cluster. This is why we use
esi_cluster_setup() to set up a parallel computing client.
They are perfectly reproducible on any other cluster or local machine
by instead using slurm_cluster_setup() or local_cluster_setup()
respectively.
The following Python code demonstrates how to use ACME to perform a parallel bootstrap of the classification accuracy of three different scikit-learn classifiers.
We start by loading the wine dataset
from scikit-learn and splitting it into training and testing sets using the
train_test_split() function from sklearn.model_selection.
from sklearn import datasets
from sklearn.model_selection import train_test_split
data = datasets.load_wine()
X_train, X_test, y_train, y_test = train_test_split(data.data, data.target, test_size=0.5, random_state=42)
Training and Evaluating The Classifiers
Next, we define three scikit-learn classifiers: K nearest neighbors
(KNeighborsClassifier), a neural network model
(MLPClassifier), and a support vector machine
(SVC). We train each of them on the training set and
evaluate their accuracy on the test set using the respective score
methods.
from sklearn.neighbors import KNeighborsClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.svm import SVC
neighbors = KNeighborsClassifier(n_neighbors=3)
NeuralNet = MLPClassifier(random_state=1, max_iter=300)
SVM = SVC(gamma='auto')
neighbors.fit(X_train, y_train)
NeuralNet.fit(X_train, y_train)
SVM.fit(X_train, y_train)
print(f"K nearest neighbors accuracy: {neighbors.score(X_test, y_test):.3f}")
print(f"Naive Bayes accuracy: {NeuralNet.score(X_test, y_test):.3f}")
print(f"Support Vector Machine accuracy: {SVM.score(X_test, y_test):.3f}")
Because we evaluated the accuracy on the test data, we only get one accuracy measure per classifier. However, we would like to have a distribution of accuracies to later compare the confidence intervals. To achieve this, we can use bootstrapping.
Bootstrapping and Confidence Intervals
We define a function bootstrap_model_accuracy that resamples the test
set with replacement and calculates the accuracy of each classifier on
the resampled data. We will use ACME to parallelize the bootstrapping
process for efficiency.
from sklearn.utils import resample
from acme import cluster_cleanup, esi_cluster_setup, ParallelMap
import numpy as np
def bootstrap_model_accuracy(X_test, y_test, seed):
X_resamp, y_resamp = resample(X_test, y_test, replace=True)
return SVM.score(X_resamp, y_resamp), NeuralNet.score(X_resamp, y_resamp), neighbors.score(X_resamp, y_resamp)
client = esi_cluster_setup(partition="8GBXS", n_workers=10)
nboot = 100
seeds = np.linspace(0, nboot, nboot, dtype=int)
with ParallelMap(bootstrap_model_accuracy, X_test, y_test, seeds, n_inputs=nboot, write_worker_results=False,result_shape=(None,3)) as pmap:
results = pmap.compute()
cluster_cleanup(client) # close the cluster if you don't need it anymore
We now have a distribution of accuracies for each classifier. This means we can calculate a confidence interval for each classifier.
from scipy.stats import sem, t
def CInt(data, confidence=0.95):
n = len(data)
m = np.mean(data)
std_err = sem(data)
h = std_err * t.ppf((1 + confidence) / 2, n - 1)
return m - h, m + h
print(f"K nearest neighbors CI:{CInt(results[:, 2])[0]:.3f} to {CInt(results[:, 2])[1]:.3f}")
print(f"NeuralNet CI: {CInt(results[:, 1])[0]:.3f} to {CInt(results[:, 1])[1]:.3f}")
print(f"Support Vector Machine CI: {CInt(results[:, 0])[0]:.3f} to {CInt(results[:, 0])[1]:.3f}")
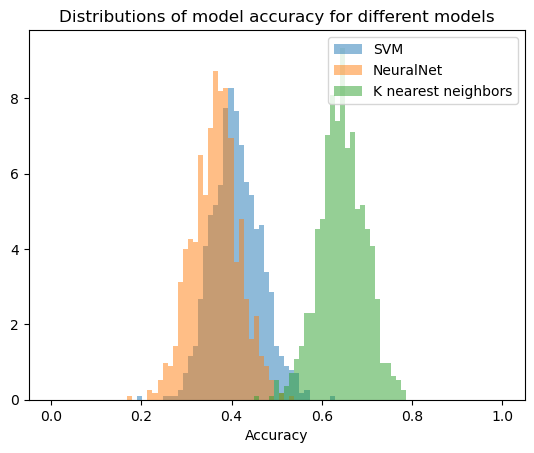
We can now go ahead and also plot our bootstrapped results as histograms.
# define bin edges for the histogram
bins = np.linspace(0,1,90)
# plot the distribution of the scores
plt.hist(results[:,0],bins=bins,alpha=0.5,label="SVM",density=True)
plt.hist(results[:,1],bins=bins,alpha=0.5,label="NeuralNet",density=True)
plt.hist(results[:,2],bins=bins,alpha=0.5,label="K nearest neighbors",density=True)
plt.xlabel("Accuracy")
plt.title("Distributions of model accuracy for different models")
plt.legend()
This is a simple procedure to compare the performance of different classifiers and we could have also achieved the same using a for loop. However, the advantage of using ACME becomes apparent when we are using larger data sets and more complex models. In this case, the bootstrapping process can take a long time and parallelization is necessary to speed up the process. ACME allows us to parallelize the bootstrapping process with just a few lines of code.
