Functional Connectivity Tutorial
Note
These examples were run on the ESI HPC cluster. This is why we use
esi_cluster_setup() to set up a parallel computing client.
They are perfectly reproducible on any other cluster or local machine
by instead using slurm_cluster_setup() or local_cluster_setup()
respectively.
The following Python code demonstrates how to use ACME to concurrently compute the functional connectivity in fMRI datasets of multiple subjects.
We start by downloading a publicly available fMRI dataset comprising BOLD signals from 33 adults using the Nilearn package:
from nilearn import datasets
data = datasets.fetch_development_fmri(age_group="adult")
The brain regions of interest (ROI) are taken from Power et al. 2011 comprising an atlas of 264 areas.
from nilearn import plotting
atlas = datasets.fetch_coords_power_2011(legacy_format=False)
Next, we extract ROI-averaged time-series from the downloaded fMRI data. To do this, we first use the MNI coordinates of every ROI in the atlas as seeds for masking the fMRI data:
from nilearn.maskers import NiftiSpheresMasker
atlasCoords = np.vstack((atlas.rois['x'],
atlas.rois['y'],
atlas.rois['z'])).T
masker = NiftiSpheresMasker(seeds=atlasCoords, smoothing_fwhm=6,
radius=5., detrend=True,
standardize=True,
low_pass=0.1, high_pass=0.01, t_r=2)
Next, we pick a random subject (Number 0) and perform the signal extraction removing nuisance regressors shipped with the fMRI dataset
timeseries = masker.fit_transform(data.func[0],
confounds=data.confounds[0])
The extracted BOLD time-courses are contained in a (#time-points x #ROIs) = (168 x 264) array.
We choose partial correlation
as statistical dependence metric for computing the functional connectivity
across all 264 regions in our atlas.
Specifically, we compute the sparse inverse covariance matrix of the BOLD
data with a cross-validated choice of the \(\ell^1\)-penalty using
scikit-learn’s GraphicalLassoCV
from sklearn.covariance import GraphicalLassoCV
estimator = GraphicalLassoCV()
estimator.fit(timeseries)
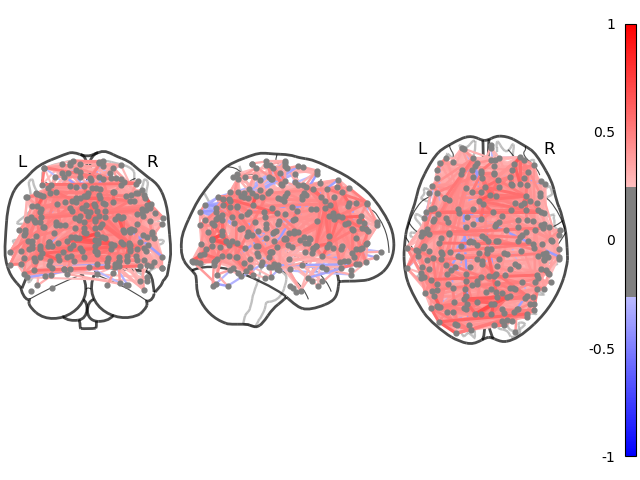
To visualize the computed inter-areal functional connectivity of Subject #0,
we use Nilearns plot_connectome()
from nilearn import plotting
plotting.plot_connectome(estimator.covariance_, atlasCoords,
title=f"Subject #{subIdx}",
edge_threshold="95%", node_size=20,
colorbar=True, edge_vmin=-1, edge_vmax=1,
figure=fig)
Functional Connectivity in Parallel
In order to compute the functional connectome of multiple subjects concurrently,
we first capsulate the computational steps in a Python function called
compute_connectome which we define inside a dedicated module connectome.py:
import numpy as np
from nilearn import datasets
from nilearn.maskers import NiftiSpheresMasker
from sklearn.covariance import GraphicalLassoCV
from numpy.typing import NDArray
# Format atlas coordinates
atlas = datasets.fetch_coords_power_2011(legacy_format=False)
atlasCoords = np.vstack((atlas.rois['x'],
atlas.rois['y'],
atlas.rois['z'])).T
def compute_connectome(subidx : int) -> NDArray[np.float64]:
"""
Compute functional connectome of single subject
Parameters
----------
subidx : int
Subject number
Returns
-------
con : 2D np.ndarray
Functional connectivity matrix
"""
# Take stock of data on disk
data = datasets.fetch_development_fmri(age_group="adult")
# Extract fMRI time-series averaged within spheres @ atlas coords
masker = NiftiSpheresMasker(seeds=atlasCoords, smoothing_fwhm=6,
radius=5., detrend=True,
standardize=True, low_pass=0.1,
high_pass=0.01, t_r=2)
timeseries = masker.fit_transform(data.func[subidx],
confounds=data.confounds[subidx])
# Compute functional connectivity b/w brain regions
estimator = GraphicalLassoCV()
estimator.fit(timeseries)
return estimator.covariance_
We now define a set of subjects to analyze and set up a parallel computing client to do the actual processing
from connectome import compute_connectome
from acme import esi_cluster_setup, ParallelMap
subjectList = list(set(range(20)).difference([7, 8, 11, 12, 19]))
myClient = esi_cluster_setup(n_workers=8, mem_per_worker="8GB",
cores_per_worker=2, partition="E880")
The return value of compute_connectome is always a
(#ROIs x #ROIs) = (264 x 264) matrix for all subjects. Thus, we
want to store the computed functional connectivity matrices in a single
shared 3d-array for easier post-processing. We use ParallelMap’s
result_shape keyword for that:
with ParallelMap(compute_connectome,
subjectList,
result_shape=(264, 264, None)) as pmap:
pmap.compute()
The resulting array can be accessed like any other HDF5 dataset, e.g., to visualize the inter-areal functional connectivity of Subject #4, we can use
import h5py
sub4 = h5py.File(pmap.results_container, "r")["result_3"][()]
plotting.plot_connectome(estimator.covariance_, atlasCoords,
title=f"Subject #{subIdx}",
edge_threshold="95%", node_size=20,
colorbar=True, edge_vmin=-1, edge_vmax=1,
figure=fig)
For more information about using result_shape and virtual HDF5 datasets,
see Collect Results in Single Dataset
